Fruit-fly inspired neighbor encoding for classification
Intro: Biological systems (such a neural networks [4,5] convolutions [7], dropout [3], attention mechanisms [6,9]) have served as inspiration to modern deep learning systems, demonstrating amazing empirical performance in areas of computer vision, natural language programming and reinforcement learning. Such learning systems are not biologically viable anymore, but the biological inspirations were critical. This has motivated a lot of research into identifying other biological systems that can inspire development of new and powerful learning mechanisms or provide novel critical insights into the workings of intelligent systems. Such neurobiological mechanisms have been identified in the olfactory circuit of the brain in a common fruit-fly, and have been re-used for common learning problems such as similarity search [2,10], outlierdetection [1] and word embedding [8].
More precisely, in the fruit-fly olfactory circuit, an odor activates a small set of Kenyon Cells (KC) which represent a ``tag'' for the odor. This tag generation process can be viewed as a natural hashing scheme [2] termed FlyHash, which generates a high dimensional but very sparse representation (2000 dimensions with 95\% sparsity). This tag/hash creates a response in a specific mushroom body output neuron (MBON) -- the MBON-α'3 -- corresponding to the perceived novelty of the odor. [1] ``interpret the KCàMBON-α'3 synapses as a Bloom Filter''that creates a ``memory'' of all the odors encountered by the fruit-fly, and reprogram this Fly Bloom Filter (FBF) as a novelty detection mechanism that performs better than other locality sensitive Bloom Filter-based novelty detectors for neural activity and vision datasets.
This “learning” of the data distribution (for the purposes of novelty detection) has some interesting dynamics. First, the FBF encodes the data distribution in a single-pass manner without requiring to visit the same example twice – the relevant information for an example has been “stored” in the FBF– surfacing two advantages: (i) once processed, there is no need to retain an example in memory, allowing the encoding without much memory overhead, (ii) thismechanism allows the FBF encoding to happen in an online manner. The single-pass, online learning is critical in many situations such as (a) the learning happens in low-memory “edge” devices where one cannot retain a training set to repeatedly revisit, or (b) the examples have to be discarded after a short amount of time due to privacy concerns. There are situations where both reasons are valid – models need to be regularly updated with new data, but the retraining cannot access old data due to privacy regulations.
Additionally, FBF learning can be accomplished solely with additions, ORs and sort operations without requiring complex novel computer hardware, mathematical operators and multiple epochs over the training examples. To take advantage of these, the primary motivation of this project is to extend the usage of FBF to the supervised learning set-up. In particular, the goals of this project are: (a) to devise a supervised classification scheme based on the simple learning dynamics of the FBF (b) empirically validate whether such a supervised classification scheme is useful and competitive when learning needs to happen with a single pass, and (c) establish generalization guarantees for such a learner.

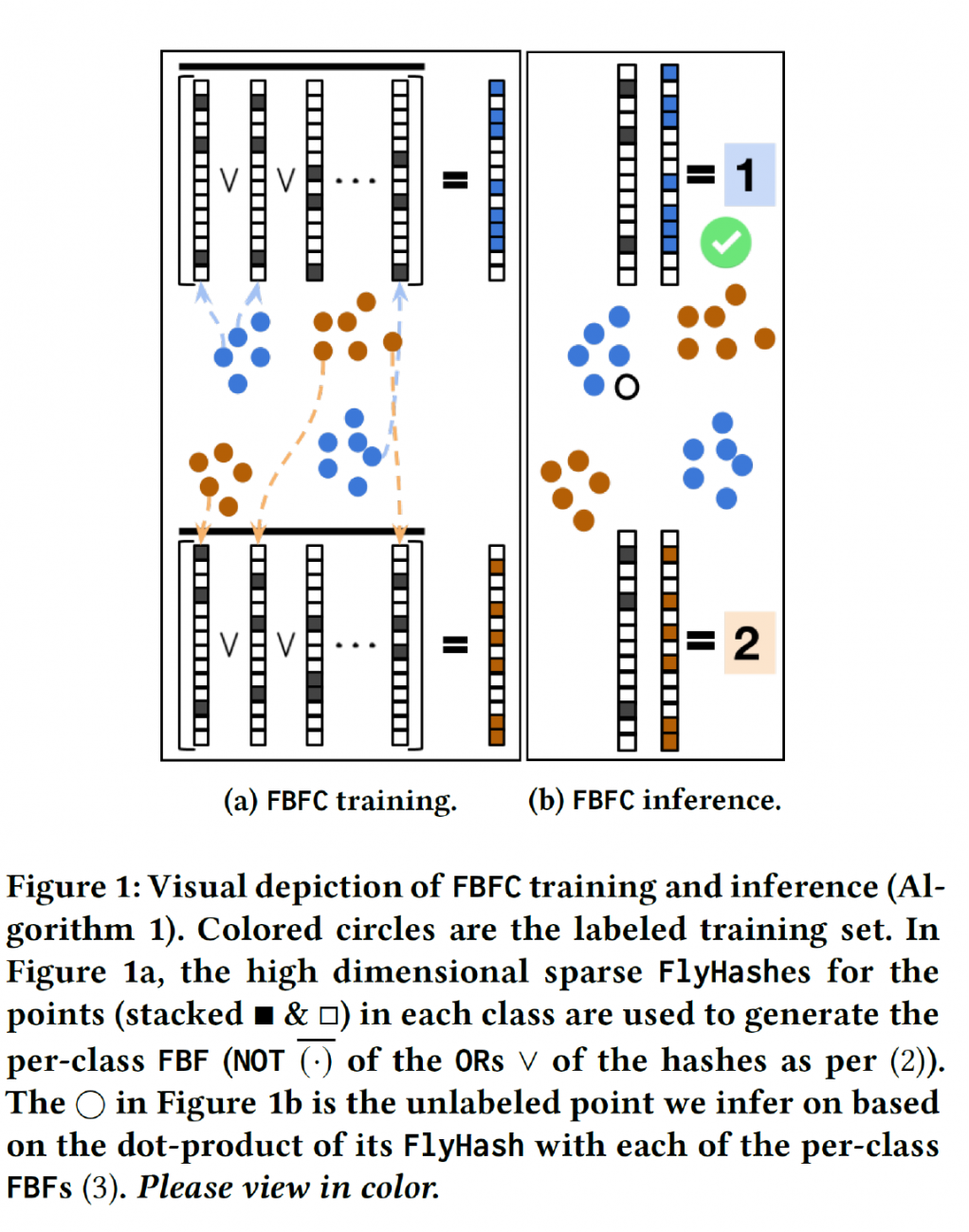
Results: We developed a new classifier that effectively encodes the different local neighborhoods for each class with a per-class Fly Bloom Filter. The inference on test data requires an efficient FlyHash operation followed by a high-dimensional but very sparse, dot product with the per-class Bloom Filters. On the theoretical side, we establish conditions under which the predictions of our proposed classifier agrees with the predictions of the nearest neighbor classifier. We extensively evaluate our proposed scheme with 71 data sets of varied data dimensionality to demonstrate that the predictive performance of our proposed neuroscience inspired classifier is competitive to the nearest-neighbor classifiers and other single-pass classifiers.
Acknowledgement: This research has been supported by the NSF IFML institute.
References
[1] Sanjoy Dasgupta, Timothy C Sheehan, Charles F Stevens, and Saket Navlakha. A neural data structure for novelty detection. Proceedings of the National Academy of Sciences, 115(51):13093 -13098, 2018.
[2] Sanjoy Dasgupta, Charles F Stevens, and Saket Navlakha. A neural algorithm for a fundamental computing problem. Science, 358(6364):793 - 796, 2017.
[3] Geo_rey E Hinton, Nitish Srivastava, Alex Krizhevsky, Ilya Sutskever, and Ruslan R Salakhutdinov. Improving neural networks by preventing co-adaptation of feature detectors. arXiv preprint arXiv:1207.0580, 2012.
[4] Koray Kavukcuoglu, Pierre Sermanet, Y-Lan Boureau, Karol Gregor, Michaël Mathieu, and Yann L Cun, Learning convolutional feature hierarchies for visual recognition. In Advances in neural information processing systems, pages 1090 - 1098, 2010.
[5] Alex Krizhevsky, Ilya Sutskever, and Geo_rey E Hinton. Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems, pages 1097 -1105, 2012.
[6] Hugo Larochelle and Geo_rey E Hinton. Learning to combine foveal glimpses with a third-order Boltzmann machine. In Advances in neural information processing systems, pages 1243_1251, 2010.
[7] Yann Lecun and Yoshua Bengio. Convolutional Networks for Images, Speech and Time Series, pages
255_258. The MIT Press, 1995.
[8] Yuchen Liang, Chaitanya K Ryali, Benjamin Hoover, Leopold Grinberg, Saket Navlakha, Mohammed J Zaki, and Dmitry Krotov. Can a fruit _y learn word embeddings? arXiv preprint arXiv:2101.06887, 2021.
[9] Volodymyr Mnih, Nicolas Heess, Alex Graves, et al. Recurrent models of visual attention. In Advances in neural information processing systems, pages 2204 - 2212, 2014.
[10] Chaitanya K Ryali, John J Hop_eld, Leopold Grinberg, and Dmitry Krotov. Bio-inspired hashing for unsupervised similarity search. arXiv preprint arXiv:2001.04907, 2020.