Robust MRI Using New Algorithms for Generative Machine Learning
Magnetic Resonance Imaging (MRI) is currently expensive and time consuming. Exam times can take upwards of 30 minutes to an hour. As a result, patient motion can cause substantial image artifacts and patient comfort becomes an issue. Our research is focused on novel algorithms that give state of the art applications for accelerating MR imaging, estimating patient motion, and improving motion correction. We have begun validating these algorithms in collaboration with clinicians at Dell Medical School. Furthermore, we have identified systematic biases in how public MR datasets are used for assessing machine learning algorithms leading to overly optimistic results. We have released code to reproduce the results of our papers, as well as software repositories to make it easier for users to train models on other imaging inverse problems.
Foundational Innovation
The use of end-to-end deep learning methods is hindered by their black-box nature, due to lack of interpretability, trust, and robustness guarantees. Model-based deep learning is the general idea of integrating knowledge about the problem in both the network architecture and the training process. We have been arguing for model-based methods [Shlezinger-PIEEE23], showing how they lead to architectures that are better understood, can be debugged during failures, require fewer data to train, and provide uncertainty estimates compared to their black-box counterparts.
Our foundational innovation has been the separation of the image prior (trained as a powerful diffusion generative model) with the physics of data acquisition (performed by Langevin dynamics during inference). We trained the first successful diffusion model for generating high-quality MR images from the Facebook FastMRI dataset and we developed the Langevin inference algorithm for reconstructing from sub-sampled multi-coil MR measurements [Jalal-NeurIPS21]. We proved that our algorithm is robust to natural shifts in the measurement operator and the imaging anatomy.
Our theoretical results establish bounds on the quality of the image reconstruction that depend on the complexity of the data distribution. We defined a novel measure of metric entropy and proved it characterizes the number of measurements needed to achieve good reconstruction quality. We have also characterized and derived a bound on the reconstruction error when the training and test set distributions differ. Our algorithm and theoretical guarantees were built on foundational work on sampling and compressed sensing performed in the IFML institute.
Novel Algorithm: Posterior Sampling from pre-trained Generative models
A significant body of prior work focuses on developing end-to-end deep learning methods that reconstruct images from MR measurements. Such methods perform extremely well when evaluated in-distribution, but are fragile to changes to anatomy and scanner settings during inference [Jalal-NeurIPS21,Shimron-PNAS22]. The reason for this fragility is the explicit coupling of the measurement model and the image prior during training. In essence, a prescribed measurement operator and a dataset must both be specified at train time. When the test-time conditions differ, then the reconstructions will suffer from artifacts due to generalization error. In practice, clinical MRI demands the flexibility to change measurement settings on a per-patient basis in order to accommodate the natural heterogeneity in patient populations.
To overcome this limitation, we explicitly decouple the measurement model and the image prior. We use diffusion generative models to learn image priors without reference to the measurement process. We use established and principled statistical physical models to separately describe the likelihood function of the measurement system. We then pose the image reconstruction task as sampling from the posterior distribution. As a result, we are able to accelerate the MRI scan by factors of 4x-12x while maintaining high image quality. As shown in Figure 1, we are able to match the quality of state of the art end-to-end methods in-distribution, while also handling test-time distribution shifts such as sub-sampling rate and imaging anatomy. A benefit of this approach is the ability to run the algorithm multiple times in parallel and thus sample multiple times from the posterior. This allows us to estimate uncertainty in the reconstruction (Figure 2), potentially providing more nuanced information about the quality of the reconstruction to the clinician.
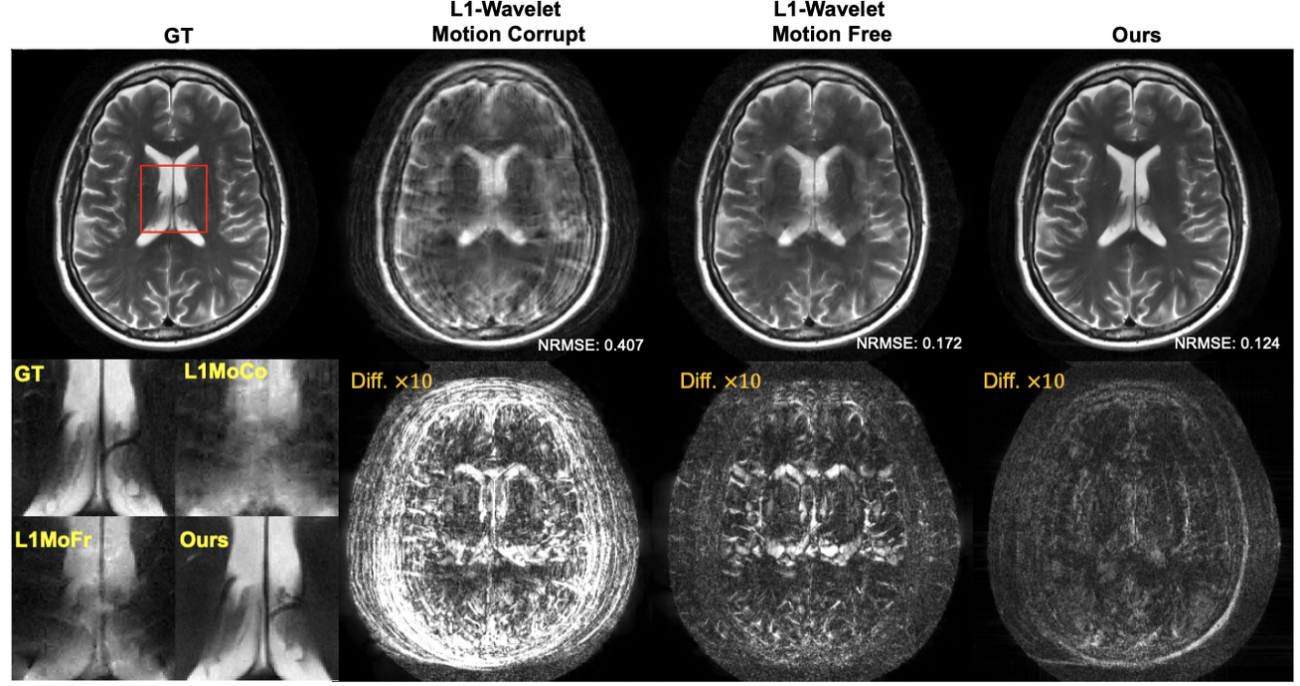
In addition to estimating uncertainty in the reconstruction, our framework also allows us to incorporate uncertainty in the measurement model itself. For example, patient motion during the scan leads to ambiguity in the acquired measurements. By modeling the motion as an unknown random variable, we can extend the reconstruction to sample from the joint posterior of both the image and the motion parameters [Levac-ISBI23], as shown in Figure 3. In contrast, end-to-end methods are not able to anticipate all possible motion patterns due to the nature of the training approach [Levac-MICCAI22].
We have successfully applied our framework to prospectively accelerated scans of healthy volunteers with informed consent and IRB approval at the UT Dell Medical School [Arvinte-ISMRM22]. While our preliminary results are promising, our proposed methods still require external validation on patients in collaboration with clinicians. In addition, several technical barriers to successful clinical translation remain; e.g., reconstruction speed and nonlinear physical model. These practical problems have now fed back into and guided our foundational work exploring these topics, with the goal of advancing clinical translation.
Implicit Data Crimes
Work led by and in collaboration with Dr. Efrat Shimron, Mr. Ke Wang, and Prof. Michael Lustig, UC Berkeley EECS Department.
Many of the advances in machine learning have been enabled by the availability of large and open databases for training. In recent years, large medical imaging datasets have also been released to help push the frontier of medical AI. However, these datasets were intended for downstream medical imaging applications, for example for automated diagnosis, segmentation, or image quality assessment. We have shown that when these data are used for MRI reconstruction, they can lead to biased, overly optimistic results [Shimron-PNAS22]. The reason for this is that the MR data are often processed following the reconstruction – for example, with interpolation, intensity correction, and image compression. These processing steps introduce systematic changes to the data that machine learning algorithms can use to artificially improve performance. Surprisingly, the use of processed data may lead to good-looking results with low errors. However, these results are overly optimistic due to the effect of the hidden data processing pipelines, and they are not actually attainable in practice for raw data. We coined the term “implicit data crimes” to indicate that “off-label” use of imaging databases could lead to bias in AI algorithms. Figure 4 demonstrates this effect: when an AI algorithm that is trained on processed data is also tested on processed data, the result is optimistically good. However, if it is applied to non-processed data directly from the MRI scanner, the reconstruction quality degrades, and pathology is no longer visible.
Figure 1. Fully sampled MR images (left column) compared to state of the art end-to-end reconstruction methods (MoDL and E2E-Varnet) and our proposed Langevin sampling algorithm. Row 1: In-distribution performance of all three algorithms is comparable. Row 2: At 12x acceleration, end-to-end methods fail. Row 3: When the imaging anatomy differs, end-to-end methods fail.
Figure 2. Our framework allows for sampling from the posterior distribution (left). Multiple samples can be drawn and used to form the conditional expectation (left-middle). The samples can also be used to evaluate uncertainty (right-middle), which closely matches with the true error (right).
Figure 3. Comparison of reconstruction methods in the presence of motion. Left: fully-sampled reference image. Left-middle: Compressed sensing reconstruction that does not account for motion during the scan. Right-middle: Compressed sensing reconstruction when no motion occurs during the scan. Right: Our Langevin sampling reconstruction which simultaneously estimates the unknown motion. The bottom row shows a zoom-in of the ventricles as well as the absolute difference between reference and each reconstruction (amplified by 10x).
Figure 4. On the left is a fully sampled, “gold standard” MRI scan of a knee requiring a long scan time. The middle column shows the result of an AI algorithm applied to a simulated fast scan from processed, open-source data. The right column shows the result of applying the AI algorithm on raw, non-processed data. There is a considerable loss of detail, in particular in the central region of the knee consisting of menisci, cartilage, and ligaments. Here, pathology manifests as subtle changes in signal intensity such as the faint dark horizontal line indicated by the yellow arrow in the top panel. The reconstructed image from processed data preserves these subtle features, while the reconstruction of non-processed data obscures the pathology.
References
[Arvinte-ISMRM22] M. Arvinte, A. Jalal G. Daras, E. Price, A.G. Dimakis, J.I. Tamir, “Single-Shot Adaptation using Score-Based Models for MRI Reconstruction.” ISMRM Annual Meeting, 2022.
[Levac-MICCAI22] B.R. Levac, S. Kumar, S. Kardonik, and J.I. Tamir, “FSE Compensated Motion Correction for MRI Using Data Driven Methods.” 25th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2022.
[Levac-ISBI23] B.R. Levac, A. Jalal, and J.I. Tamir, “Accelerated Motion Correction for MRI using Score-Based Generative Models.” IEEE International Symposium on Biomedical Imaging (ISBI), to appear, 2023.
[Jalal-NeurIPS21] A. Jalal, M. Arvinte, G. Daras, E. Price, A.G. Dimakis, and J. Tamir. "Robust compressed sensing mri with deep generative priors." Advances in Neural Information Processing Systems (NeurIPS) 2021.
[Shlezinger-PIEEE23] N. Shlezinger, J. Whang, Y.C. Eldar, and A.G. Dimakis, "Model-Based Deep Learning," Proceedings of the IEEE, to appear, 2023.
[Shimron-PNAS22] E. Shimron, J.I. Tamir, K. Wang, and M. Lustig. “Implicit data crimes: Machine learning bias arising from misuse of public data.” Proceedings of the National Academy of Sciences (PNAS), 2022.
Source Code
- CSGM-MRI-Langevin: Source code to reproduce the results in [Jalal-NeurIPS21]
- Nufft_Torch: Library for non-uniform MRI sampling, developed for [Levac-ISBI23]
- Score-Diffusion-Training: Library for training score-based generative models
- Score-Diffusion-Sampling: Library for prior and posterior sampling for arbitrary imaging inverse problems
Media Highlights
- A vital medical imaging technique is getting a major reconstruction, thanks to researchers at the University of Texas at Austin. Oracle Research in Action Podcast, Nov 9, 2022.
- ‘Off Label’ Use of Imaging Databases Could Lead to Bias in AI Algorithms, multiple outlets (including Science Daily, UC Berkeley, UT Austin Engineering, Daily Texan), March, 2022.
- How new machine learning techniques could improve MRI scans, Amazon Science, Jan. 12, 2022.