SCALP - Supervised Contrastive Learning for Cardiopulmonary Disease Classification and Localization in Chest X-rays using Patient Metadata
Research thrust(s): Use-Inspired Applications
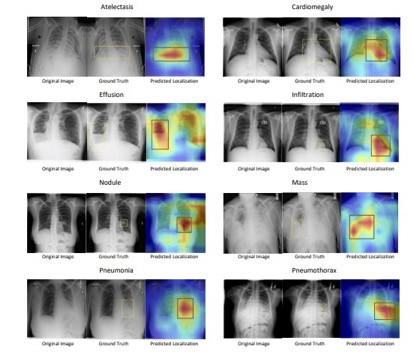
Chest X-rays (CXR) are one of the most common imaging tools used to examine cardiopulmonary diseases. Currently, CXRs diagnosis primarily relies on professional knowledge and meticulous observations of expert radiologists. Automated systems for medical image classification face several challenges. First, it heavily depends on manually annotated training data which requires highly specialized radiologists to do manual annotation. Radiologists are already overloaded with their diagnosis duties and their hourly charge is costly. Second, common data augmentation methods in computer vision, such as crop, mask, blur, and color jitter can significantly alter medical images and generate inaccurate clinical images. Third, unlike images in the general domain, there is subtle variability across medical images. In addition, a significant amount of the variance is localized in small regions. Thus, there is an unmet need for deep learning models to capture the subtle differences across diseases by attending to discriminative features present in these localized regions.
Fundamental Algorithmic Innovation: Data Augmentation and Supervised Contrastive Learning
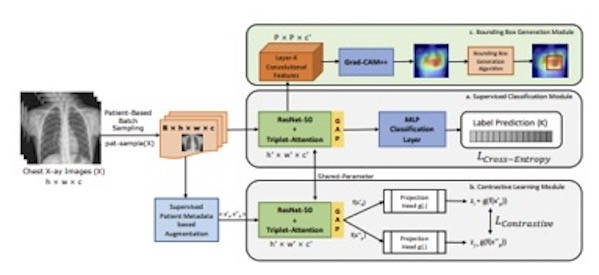
We propose a novel and simple data augmentation method based on patient metadata and supervised knowledge such as disease labels to create clinically accurate positive samples for chest X-rays. The supervised classification loss helps SCALP create decision boundaries across different diseases while the patient-based contrastive loss helps SCALP learn discriminative features across different patients. Compared to other baselines, SCALP uses simpler ResNet-50 architecture and performs significantly better. SCALP uses GradCAM++ to generate activation maps that indicate the spatial location of the cardiopulmonary diseases.
Research highlights:
- Augmentation technique for contrastive learning that utilizes both patient metadata and supervised disease labels to generate clinically accurate positive and negative keys. Positive keys are generated by taking two chest radiographs of the same patient P while negative keys are generated using radiographs from patients other than P and having the same disease as P.
- Novel unified framework to simultaneously improve cardiopulmonary disease classification and localization. We go beyond the conventional two-staged training (pretraining and fine-tuning) involved in contrastive learning. We demonstrate that single-staged end-to-end supervised contrastive learning can improve existing baselines significantly.
- We propose an innovative rectangular Bounding Box generation algorithm using pixel-thresholding and dynamic programming.
We propose a simple and effective end-to-end framework SCALP using supervised contrastive learning to identify cardiopulmonary diseases in chest X-ray. We go beyond two-stage training (pre-training and fine-tuning), and demonstrate that an end-to-end supervised contrastive training using two images from the same patient as a positive pair, can significantly outperform SOTA on disease classification. SCALP can jointly model disease identification and localization using the linear combination of contrastive and classification loss. We also propose a time-efficient Bounding Box generation algorithm that generates bounding boxes from the attention map of SCALP. Our extensive qualitative and quantitative results demonstrate the effectiveness of SCALP and its state-of-the-art performance.
Jaiswal, A.; Li, H., Zander, C., Han, Y., Rousseau, J., Peng, Y., & Ding, Y. (2021). SCALP - Supervised Contrastive Learning for Cardiopulmonary Disease Classification and Localization in Chest X-rays using Patient Metadata. IEEE International Conference on Data Mining (ICDM 2021), Dec 7-10, 2021, Auckland, New Zealand.
This research has been supported by the NSF AI Institute for Foundations of Machine Learning (IFML), Amazon Machine Learning Research Award. It also was supported by the National Library of Medicine under Award No. 4R00LM013001.